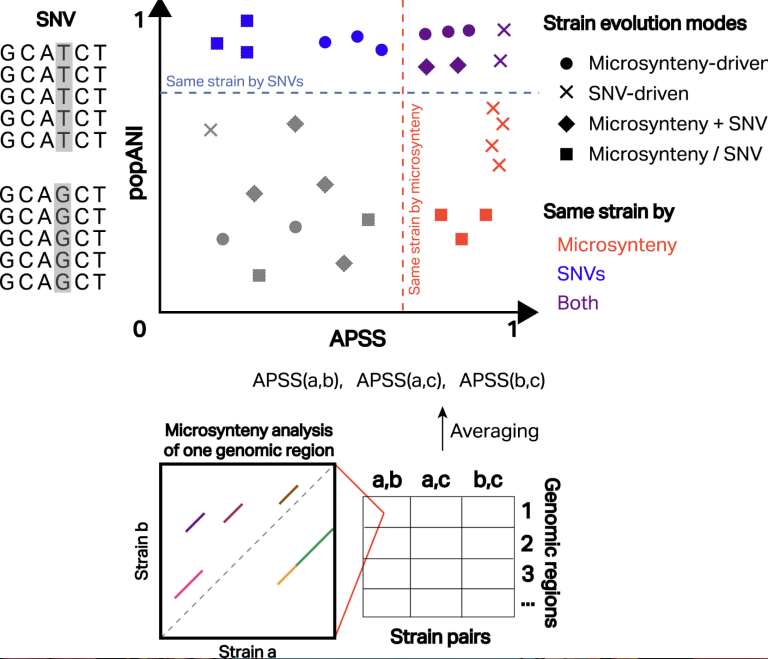
We were happy to see Hagay Enav’s Nature Biotechnology paper introducing the use of microsynteny for strain comparisons with the development of the tool SynTracker featured in Cell Reports Methods. Peiwan Cai and Tal Koren from the Columbia University Irving Medical Center in NY, write in their article entitled “Microsynteny is a powerful front for microbial strain tracking“:
“Overall, SynTracker is a robust and scalable microsynteny-based strain-tracking tool for metagenomic data. Moving beyond reference marker genes and requiring only a fraction of the genome, it has the flexibility to work with microbial elements that have relatively lower sequencing coverage, such as phages and plasmids. This can help researchers tap into less-studied microbial domains. Combining SynTracker with an SNV-based strain-inference or strain-tracking tool at a similar scope can even allow discovery of diverse strain patterns, providing researchers with a closer look at the evolutionary dynamics in microbial communities. This exciting front in strain inference and tracking is likely to fuel new discoveries in microbiome research.”