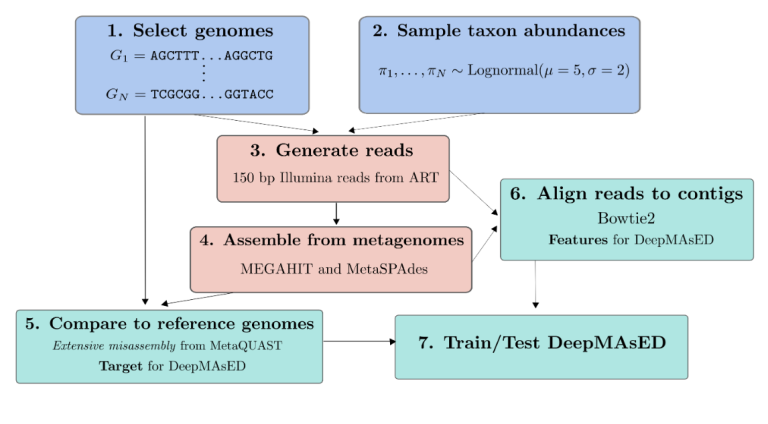
We are proud to annouce the two latest preprints from the Ley Lab, this time courtesy of the bioinformatics arm of the lab, which created two tools for the analysis of sequnece data from metagenomes. The first one, DeepMAsED, was developed in collaboration with the Department of Empirical Inference at the MPI for Intelligent Systems. DeepMAsED uses deep learning methods to detect missassebled regions in metagenomic-assembled genomes without the need for reference genomes, which allows to better evaluate the accuracy of a wide range of metagenome assembly projects. The second tool, Struo, is a pipeline for the retrieval of genomes from public repositories and the construction of custom databases for metagenome profilers, allowing the incorporation of novel genomes into the analysis of microbial communities.
The code of both tools is freely available from GitHub at
https://github.com/leylabmpi/DeepMAsED and
https://github.com/leylabmpi/Struo