Our new pre-print is up on bioRxiv! Led by Nicholas Youngblut, this work presents ResMiCo, a powerful tool for optimising metagenome assemblies.
Metagenome assembly is increasing in popularity due to decreasing Illumina sequencing costs and easier library preparation methods. Unfortunately, assembling genomes from complex metagenomes can result in misassemblies that can affect all downstream analyses. As the saying goes: “garbage in, garbage out”.
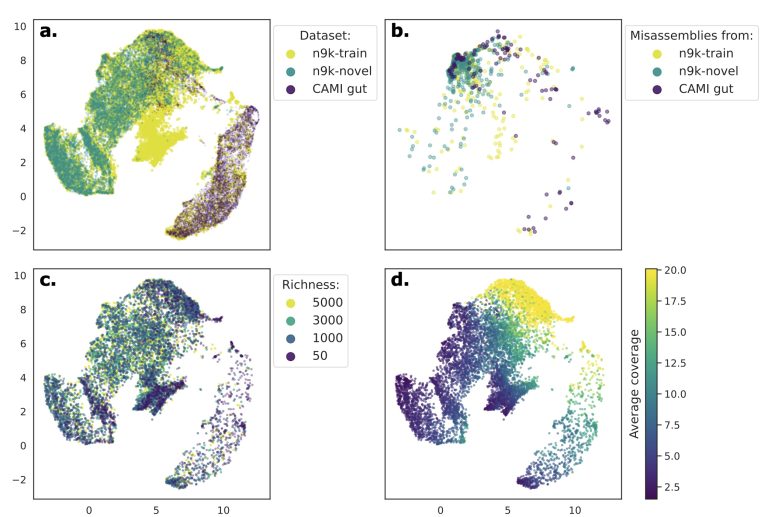
To address this issue, the Ley Lab collaborated with the labs of Gunnar Rätsch (ETH Zürich) and Bernhard Schölkopf (MPI for Intelligent Systems) to develop ResMiCo, a new deep learning (DL) approach to identify misassemblies in metagenome assemblies. ResMiCo was trained on 80x more data than the closest competitor (~2.4 Tb of compressed data). The authors show through many validations that ResMiCo performs substantially better than the state-of-the-art and generalised well to novel data. When applied to a set of real-world metagenome assemblies, ResMiCo showed that, on average, nearly 5% of contigs were misassembled.
The ResMiCo code is freely available on GitHub (https://github.com/leylabmpi/ResMiCo), includes extensive documentation, and can easily be installed via pip or conda.
ResMiCo is a general tool for accurately identifying misassemblies. Hopefully, it will be widely adopted in order to improve metagenome assemblies.
Mineeva, O., Danciu, D., Schölkopf, B., Ley, RE., Gätsch, G., Youngblut, ND. ResMiCo: increasing the quality of metagenome-assembled genomes with deep learning (2022). bioRxiv: https://doi.org/10.1101/2022.06.23.497335